During my background research, my supervisor sent me a paper for me to read, he said this paper will help me with my immunostaining protocol. The paper he sent me was conducted by researchers from Canada, Israel and Switzerland on locust foraging gene.
The background for this paper is that animals, including locusts display a astonishing genetic interaction with their environment to generate behavioral plasticity. In this study, the model animal researchers used were migratory locusts, Locusta migratoria, and desert locusts, Schistocerca gregaria. During their solitary-phase, those animals actively avoid contact with conspecifics and are relatively sedentary. However, they become more active, and start marching in bands of hoppers or flying in swarms in their gregarious phase, when their population density increases.
So far, numerous neurobiology studies have characterized the differences in locusts' sensory interneurons that faciliated this behavioral change. In contrast, little has been done to study the molecular and genetic contribution to this phenomenon.
So far, numerous neurobiology studies have characterized the differences in locusts' sensory interneurons that faciliated this behavioral change. In contrast, little has been done to study the molecular and genetic contribution to this phenomenon.
Recently, there are a handful of other well-studied animals that have shed light on the genetic and pathways that mediate environmental and behavior variations and plasticity. Recent studies have found that those foraging-related behaviors have been linked to the foraging gene (for) and a cGMP-dependent protein kinase (PKG). For example, it has been found in Drosophila melanogaster that allelic variation in for, for mRNA and PKG have been associated with a similar foraging phenomenon. Interesting, those with a rover allele (forR) exhibit a greater moving distance while feeding comparing to those with the sitter allele (forS).
In their study, researchers demonstrated that different allele variations were favoured under different environments; in a crowded population, the environment favours those with forR, while uncrowded population favours those with forS.
In their study, researchers demonstrated that different allele variations were favoured under different environments; in a crowded population, the environment favours those with forR, while uncrowded population favours those with forS.
 The researchers in this study cloned the locust for gene, placed it in an insect phylogeny of PKG, examined for expression in locust brain and measured the PKG activity. In this paper, the researchers confirmed that locusts grew up in different population-density colonies illustrated differences in development, coloration and pattern, neurophysiologies and behavior... ...
The researchers in this study cloned the locust for gene, placed it in an insect phylogeny of PKG, examined for expression in locust brain and measured the PKG activity. In this paper, the researchers confirmed that locusts grew up in different population-density colonies illustrated differences in development, coloration and pattern, neurophysiologies and behavior... ... Researchers also found that the two species of locusts, L. migratoria, and S. gregaria, shared 90% pairwire identity and a 70% pairwise similarity in for gene when comparied with D. melanogaster. Further more, the for gene in locusts had the typical PKG domains, and two cGMP-binding domains. The researchers also compared locust's for sequences with all currently known insect PKG-encoding genes. They found that within the type I PKG cluster, a group formed by the dipterans and a group with all the social insects except the wasp Vespula vulgaris can be distinguished. The two locust sequences are the locust foraging genes and are part of the PKG type I cluster.
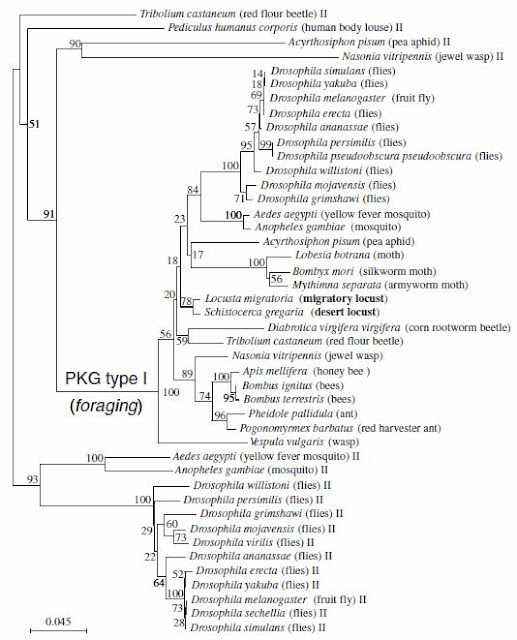 |
| This phylogenetic tree illustrated the phylogenetic analysis (Neighbour joining method) of the relationship of 44 PKG sequences spanning 30 insect species (variant sequences were discarded). Pairwise comparisons using 5,000 bootstrap replications were used to build the tree. The 2 locust species are in bold. |
During the PKG activity assay between the two phases of locusts, researchers concluded that the PKG activity in gregarious locusts was significantly higher than that in solitary locusts. The differences in PKG activity are also sex specific, with higher PKG activity found in males. However, no interaction was found between sex and phases. This sex difference in PKG activity was found for both solitary gregarious locusts.
Through neruonal staining, the researchers also identified the locust for gene expression location within the locust brain.
 |
| This group of images illustrates the FOR expression patterns in the locust brain. A: Merged confocal image of frontal 3-mm optical sections of a locust brain showing the major brain neuropils (in green, mouse antibody). B: A distinct PKG-IR region is seen as a cluster of cell bodies in the anterior midline of the brain (double staining with anti-FOR in red and mouse antibody in green). C: Similar FOR expression patterns is seen in gregarious (Gre) or solitary-reared animals (Sol), in adult male (M), female (F), and larvae. Each scale bar represents 250 mm. |
'Immunohistochemistry was as in Belay et al. (2007). Briefly, whole brains were dissected in PBS (0.1 M, pH 7.4), fixed in 4% paraformaldehyde, and blocked in 4% normal goat serum (Jackson ImmunoResearch, West Grove, PA) in 0.5% Triton X-100/PBS. The specific guinea pig antibody called anti-FOR, described in Belay et al. (2007), was used at 1:150; the neuropile marker mouse mAb nc82 was used at 1:20 (22, 23). Incubation was for 48 h at 41C. After incubation, brains were washed several times in 0.5% Triton X-100/PBS before adding a goat Cy2-conjugated anti-mouse and a Cy5-conjugated anti-guinea pig Ig (1:100, Jackson ImmunoResearch) for 24 h at 41C. For negative controls, brains were incubated in only secondary antibody, in the absence of primary antibody or in pre-absorbed anti-FOR serum. The specificity of the primary antibody, anti-FOR antibody generated in guinea pig, was measured by using Western blot immunodetection.'
What I really liked about this method section was that the steps for immunostaining was very clearly layed out, which I can copy and adjust for my experiment. The researchers also thoroughly tested the specificity of the primary antibody through Western blot immunodetection. In addition, they ensured the staining they were observing were not just background by omitting the primary antibodies in the negative control. However, the researchers did not indicate how long they fixed their tissues for, nor did they indicate the length for blocking in normal goat serum.
None the less, I got my immunohistochemistry sorted out in this paper, now it's time to fine tune the details to get everything work for my experiment!
Ref:
Lucas, C., Kornfein, R., Chakaborty, M., Schonfeld, J., Geva, N., Sokolowski, M.B., Ayali, A., 2010. The locust foraging gene. Archives of Insect Biochemistry and Physiology. 74:52-66.
Ref:
Lucas, C., Kornfein, R., Chakaborty, M., Schonfeld, J., Geva, N., Sokolowski, M.B., Ayali, A., 2010. The locust foraging gene. Archives of Insect Biochemistry and Physiology. 74:52-66.
No comments:
Post a Comment